| Version | |
| Download | 4 |
| Total Views | 181 |
| Stock | ∞ |
| File Size | 1.61 MB |
| File Type | |
| Create Date | 29/1/2024 |
| Last Updated | 22/3/2024 |
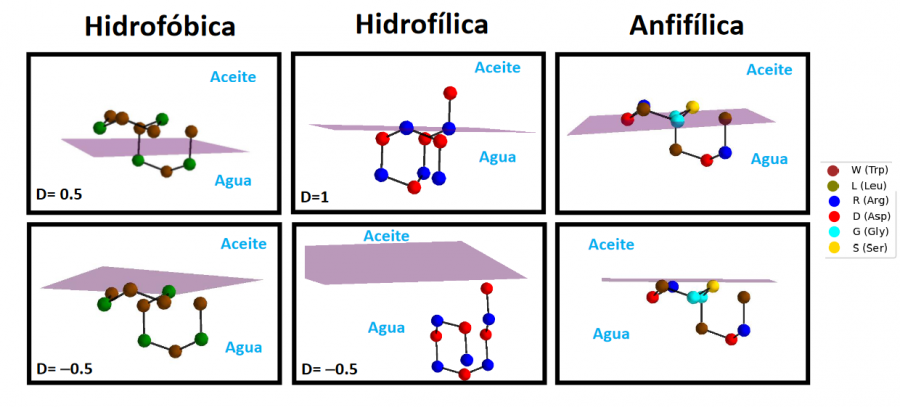
We calculated the optimal conformation for three 10-amino acid peptide sequences at interfaces with different polarity gradients. The results obtained with our approach using the quantum emulator available at CESGA.
Recently, IBM released a code aimed at solving the peptide folding problem in homogeneous environments for relatively short amino acid chains using this technology. Their approach is based on several approximations, including the representation of peptide residues using spheres that interact under the Miyazawa-Jernigan potential and the discretization of degrees of freedom. We have modified the Hamiltonian used by the IBM to include a continuous interface between a hydrophobic medium and a hydrophilic medium, thus modeling the surface of a lipid membrane. After implementing this modification in the public version of the code, we calculated the optimal conformation for three 10-amino acid peptide sequences at interfaces with different polarity gradients. The three sequences used were designed to be mainly hydrophobic, hydrophilic, and highly amphipathic, thus exhibiting different behaviors in the interface models. Results obtained with the quantum emulator available at CESGA

